Creating a Pipeline for Hypotheses Generation and Reagent Development in Alzheimer’s
By Karina Leal and Lara Mangravite
Sage Bionetworks
At the Emory-Sage-SGC TREAT-AD Center, the goal is to improve, diversify, and reinvigorate the AD drug development pipeline. How? By creating and openly distributing high-quality reagents designed to enable validation of understudied AD target hypotheses. This approach is designed to empower researchers across the field to study the understudied–and de-risk a diverse range of targets. In this post, we want to take you through our process for selecting hypotheses and target proteins that we prioritize to build high-quality reagents for target validation.
In setting up our pipeline, we focused on answering three main questions:
- How do we prioritize dark targets?
- How do we develop a target strategy for dark targets?
- How do we set up our pipeline to build TEPs and get them into research?
Overall pipeline:
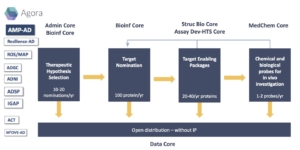
How do we prioritize dark targets?
The first step in drug discovery and development for any disease is target identification. The AD field has focused primarily on a few targets within a limited number of therapeutic hypotheses. We have set out to build a pipeline to identify not only understudied “dark” targets in AD but also understudied therapeutic pathways.
If the underlying biological mechanisms of AD are unknown, how can researchers identify targets and eventually validate and develop drugs? To answer this question, we are taking a bioinformatics analyses approach to prioritize specific genes, proteins, and pathways to generate tools for target enablement. A number of genome-scale studies have generated thousands of potential molecular targets for treating late-onset AD, including many candidate genes emanating from the Accelerating Medicine Partnership in Alzheimer’s Disease (AMP-AD) and nominated on the Agora platform. To narrow down the list of targets for therapeutic prioritization, the Bioinformatics Core has developed a gene-level scoring method. By prioritizing pathways with strong evidence as biologically relevant to AD, looking at genetic, transcriptomic, proteomic, neuropathology, and literature evidence, we have identified a number of genes to create a strategy for target enablement. We’ll go into more detail about the types of evidence used and methodology in a separate post.
How do we develop a target strategy for dark targets?
The bioinformatic analyses method provides an evidence-based strategy for prioritizing candidate genes for further development in the Center. We performed additional due diligence to develop a target strategy for these dark targets. To facilitate go/no-go decision making, we generated target dossiers. The target dossier is a complete profile that includes information gathered by an informatics approach looking at proteomic data, protein-protein interactions, cell-type expression, and a list of available reagents for each target, and a literature review, gathering available information about the hypothesized role in AD, intervention mode, and quality of available reagents. Based on this information and a feasibility assessment for potential chemical probe development, our team selected 21 targets to move forward in Year 1 and Year 2 of our project.
Our current portfolio consists of the following gene targets:
| MSN | MDK | SFRP1 |
| CD44 | SYK | PRDX1 |
| CAPN2 | FCER1G | PRDX6 |
| RABEP1 | SDC4 | GPNMB |
| C4A | NDUFS2 | CNN3 |
| SMOC1 | PLEC | SNX32 |
| STX4 | CTSH | EPHX2 |
How do we set up our pipeline to build TEPs and get them into research?
With 21 targets in hand, our Center launched a robust target-enabling package (TEP) campaign pipeline. The three technical cores–Structure Biology, Assay Development and High-Throughput Screening, and Medicinal Chemistry–are working together to build out the TEPs for each of the targets in our portfolio. Our Center is designing and developing research reagents, tools, and methods that support TEPs for prioritized targets to be used for target validation. We’ve set up target teams with representatives from each core to develop the experimental strategy for populating the different target-specific TEP components. Once the TEP components have met the quality criteria for distribution, all available data and resources, including chemical and biological probes will be distributed openly and for use without restriction. We are also engaging the community to identify potential collaborators and independent researchers to use the reagents to support target validation and mechanistic evaluation.
It’s through open science and de-risking dark targets and therapeutic hypotheses that we believe we can create an impact in accelerating drug development in AD.
